Shiny Apps
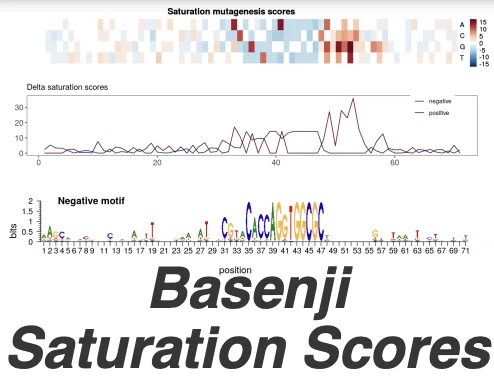
The Shiny app allows for quick visualisation of the Basenji saturation scores calculated around the 175 top pAI and top AI variants in our publication “Contextualising transcription factor binding during embryogenesis using natural sequence variation“. You can select the variants from the table on top and visualise the corresponding saturation scores.
vasa-Cas9
We provide the genotype information for the vasa-Cas9 line. This information can be used to design guide-RNAs and primers taking into account genetic variants specific to the vasa-Cas9 line. The variants were called using GATK short variant discovery pipeline. The pipeline focuses on SNPs and short INDELs. We also provide the personalized genome sequence of the vasa-Cas9 line based on the dm6 assembly. Finally, the liftOver chain files can be used to move the genomic coordinates from dm6 to vasa-Cas9 and vice-versa. The personalised genome is built using the unfiltered vcf to minimize false negative variant calls. Heterozygous variants are encoded using the IUPAC nucleotide code.
Show data
Genotype of the Drosophila vasa-Cas9 line
VCF: Genome Sequence:ReMap 2022 Drosophila coverage tracks
We provide all ReMap 2022 Drosophila tracks. The mapped bam files were provided by Fayrouz Hamal and Benoît Ballester and processed according to the ReMap analysis pipeline ( Hammal et al. 2022). According to the database annotation we were able to match 570 out of 1205 ChIP-seq samples with at least one corresponding ChIP input. All coverage tracks (ChIP samples and ChIP inputs) were generated using bamCoverage.